Research brief: Can bacteria fight back in microgravity?
Every time a human travels to space, they bring along trillions of microbial stowaways. Because of the huge impact microbes can have on human health, understanding how they are affected by microgravity is as important as understanding how the human body responds to the conditions of space.
 2019 Finalist Abinand Parthasarathy
2019 Finalist Abinand Parthasarathy
We know many of you are preparing Genes in Space proposals focused on bacteria and other microbes, so we thought we'd help you get up to speed on the latest research. We asked 2019 Finalist Abinand Parthasarathy to share one of his favorite research articles he uncovered as he was preparing his proposal on viral reactivation.
Here, Abi highlights a recent publication by a team of NASA and University of Houston researchers. Lead author Madhan Tirumalai and his colleagues asked whether microgravity influences how bacteria develop antibiotic resistance – that is, how they fight back against our efforts to kill them with antibiotics.
Guest post by 2019 Finalist Abinand Parthasarathy
What was the purpose of this study?
As we prepare for manned missions to Mars in the near future, it is important to keep astronauts safe and protected. In space, our bodies experience a host of factors, such as sleep deprivation and cosmic radiation, that make our immune systems weaker. Thus, getting viral or bacterial infections in space is especially dangerous. This study evaluates the ability of bacteria to gain antibiotic resistance (AR) in a modeled microgravity environment and tests whether the AR lasts over many generations. This will tell us more about how antibiotics can be used during long-duration missions in space.
What exactly did the researchers do?
First, the researchers grew the bacteria E.coli for a thousand generations in low-shear modeled microgravity (LSMMG). LSMMG is basically a way to simulate microgravity on Earth using a spinning container. While the bacteria were growing, they were periodically exposed to the antibiotic chloramphenicol. The researchers then tested several antibiotics on the E.coli to see whether they had developed AR. Next, they saw whether adapted E. coli can retain their AR after several generations without chloramphenicol cleaning. They also sequenced the genome of the adapted E.coli to find specific changes that might be responsible for their newly-developed AR. They compared their results to a control group of E.coli grown in LSMMG without periodic chloramphenicol exposure.
What did the researchers find?
The E. coli did show signs of AR. In fact, the E.coli not only developed resistance to chloramphenicol but also to many other antibiotics, as well! Even after the adapted E.coli were grown for several generations without chloramphenicol exposure, they still showed signs of AR. The researchers also observed twenty-two major mutations in the adapted E.coli. For example, five mutations that could underlie the observed AR were found near genes involved in drug transport and binding.
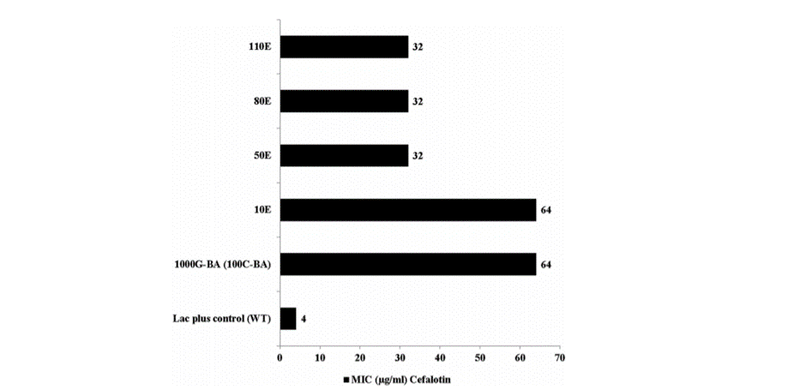
Figure 2 from Tirumalai et al. This figure shows how AR decreases after multiple generations without antibiotic exposure, but is still not lost. The number of generations is on the y-axis (1E = 1 generation) and the concentration of antibiotics necessary to affect the bacteria is on the x- axis. As the number of generations without chloramphenicol exposure increases (higher on the graph), the minimum concentration of cefalotin needed to affect the E.coli decreases – but is still higher than the concentration needed to kill E. coli that had never been exposed to chloramphenicol (the lac plus control [WT] group). The 1000G-BA is the adapted E.coli strain (0 generations without antibiotic exposure).
How does this compare with Earth?
One important observation the researchers made was that the AR responses they saw in bacteria grown in LSMMG are similar to those seen in bacteria grown in normal gravity. This means that bacteria may develop AR in a similar way in microgravity as on Earth.
What does this mean for the future of space travel?
This study shows why studying microorganisms in space is vital for future exploration. Because the containers were treated with chloramphenicol in the experiment, the E.coli were exposed to a low concentration of the antibiotics. That means even a low level of antibiotic exposure can lead to AR. This is important to know as antibiotics are sometimes used for cleaning purposes in space missions. We must be careful not to unknowingly cause AR bacteria during long-distance missions.
Inspired by this research? Have your own idea for a follow-up study? Submit it to Genes in Space! Application deadline: April 17, 2020.


