Using PCR in your Genes in Space proposal
Guest Post by Holly Christensen
Holly will be available to discuss this post and answer your questions about Genes in Space at the next #GenesInSpaceChat on Tuesday, March 20th at 8pm EDT. Submit your questions for Holly HERE.
Genes in Space invites students to research biological processes that may be altered in cosmic environments and to make use of the polymerase chain reaction (PCR) capabilities on board the International Space Station using the miniPCR machine. Once deciding what you’d like to test about DNA or RNA in space, your next step is to begin designing a PCR experiment.
Today we will discuss what goes into doing a PCR experiment. Maybe you’ve never done PCR, and that’s totally fine; a high-level understanding of PCR as a way to copy DNA or RNA is all you need to participate in Genes in Space. To get the basics, we recommend this video. However, if you’re curious and want to learn more, I hope that the article below helps you go deeper into the workings on PCR and how to set up a DNA (or RNA) amplification experiment/

Astronaut Peggy Whitson using the miniPCR aboard the International Space Station.
What is PCR
PCR is a common molecular biology technique used to make millions and millions of copies of specific pieces of DNA—we usually call that copy process DNA amplification. Whether it's a population level study or an individual gene analysis, PCR can be used to assess many aspects of DNA and RNA, mainly their presence and relative amount. PCR makes a lot of DNA, enough that we can visualize it (using gel electrophoresis), read it (sequence it), or manipulate it to our advantage (as in genetic engineering.)
PCR cycles
Each cycle of a PCR reaction has three stages: denaturation, primer annealing, and elongation.
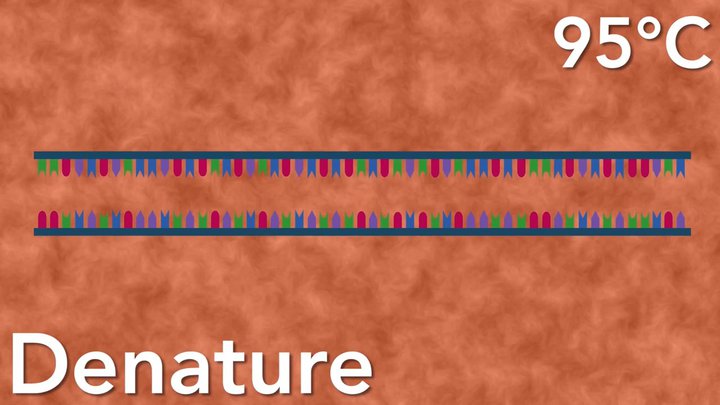
First, the denaturation of double-stranded DNA into single-stranded DNA occurs at a high temperature (94-96°C).
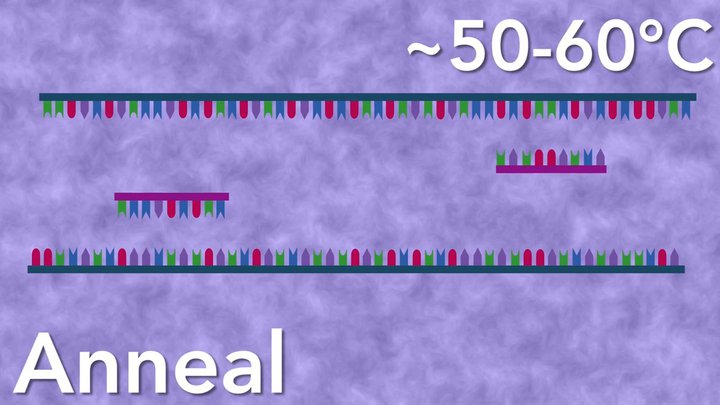
During annealing, the reaction temperature is lowered and DNA primers bind to single-stranded DNA with complementary sequences. The annealing temperature varies for each primer set and is dictated by length and sequence.
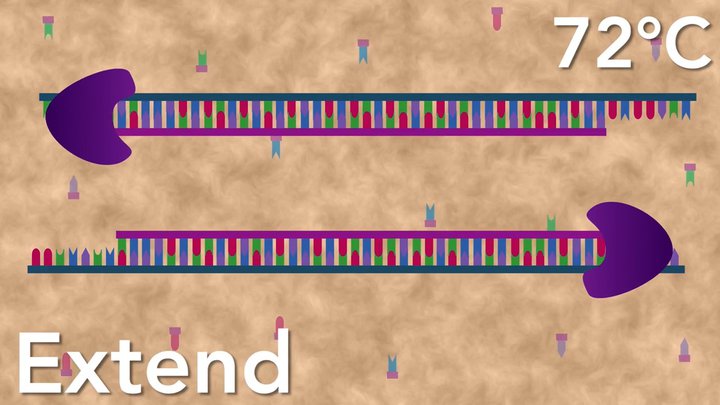
Following annealing, the temperature is raised to about 72°C for elongation by DNA polymerase. DNA polymerase uses the primers, template, and dNTPs to make new DNA strands.
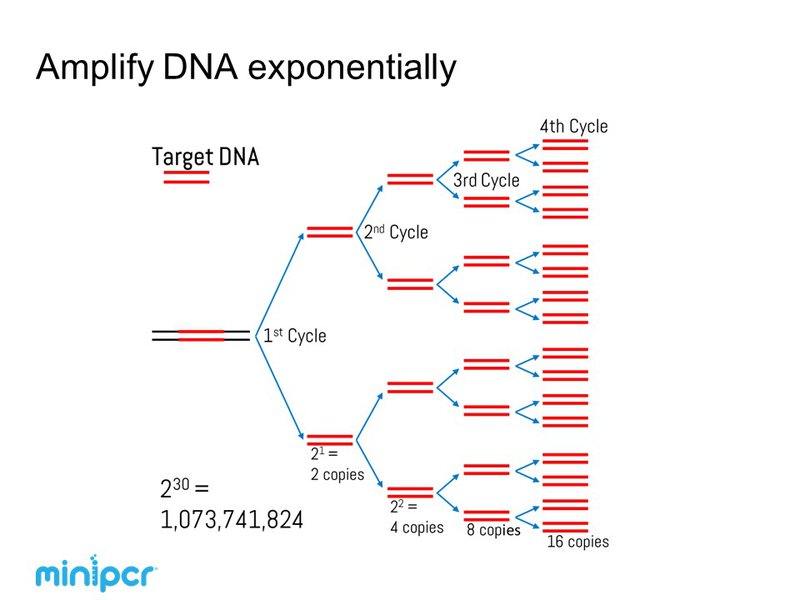
A typical PCR reaction has 20-40 cycles and can generate millions of copies of a target sequence. Initially the target sequence is only a small fraction of the template DNA. However, the primers restrict the region of synthesized DNA to the target sequence and by the end of the third cycle double-stranded PCR products containing only target sequence are present. See an animation and learn more HERE.
During the early PCR cycles, the copy number of the target sequence increases exponentially. However, as the number of PCR cycles rises, the amplification rate begins to level off. This slow down is due to the depletion of reagents, loss of polymerase activity, and competition for primer binding. Eventually, the PCR reaction will hit a plateau and no additional product will be made. For experiments exploring the relative amount of a DNA sequence across samples, the PCR reaction should end while exponential amplification is still occurring. In this instance, the number PCR cycles should be experimentally optimized for each primer pair such that the PCR product is detectable and the reaction is still in the exponential phase. Find more about this topic HERE.
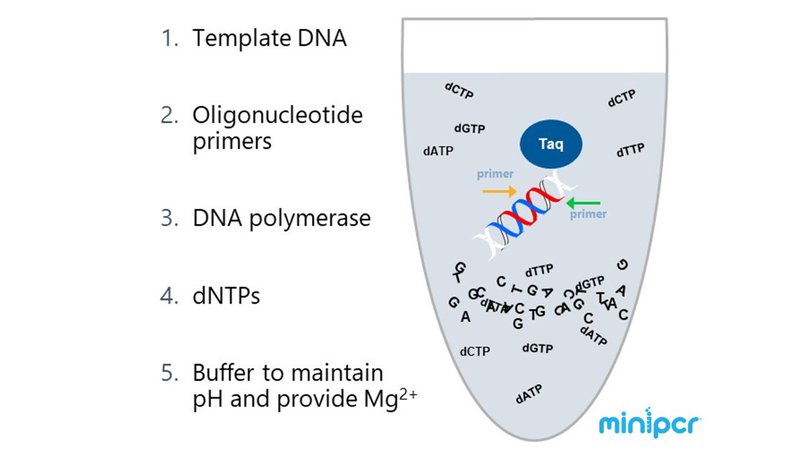
PCR Components
Template
The template is the nucleic acid that is added to the PCR reaction. Frequently, the template is DNA extracted from cells. For example, if the PCR target is on a plasmid, then plasmid DNA is added to the reaction. However, nucleic acids are also commonly modified before a PCR reaction. RNA for instance is generally converted into complementary DNA (cDNA) by reverse transcriptase before the cDNA template is amplified by PCR. As in Anna-Sophia's Genes in Space experiment, DNA can also be chemically modified prior to PCR, such as treating DNA with sodium bisulfite in order to examine DNA methylation (learn more HERE).
Primers
Oligonucleotide primers are short strands of DNA or RNA that enable DNA synthesis. To amplify a specific DNA sequence, two primers flanking that sequence are designed. However, to amplify the whole genome, many primers with random sequences would be used. Primers can also prepare PCR products for later experimental steps. For example, if the goal is to clone a PCR product into a vector following restriction enzyme digest, the desired restriction enzyme sites can be added the 5' ends of the primers.
Submitting
an application to Genes in Space does not require that you design
primers, but if you'd like to learn more about primer design, click HERE.
Thermostable DNA polymerase
DNA polymerases are the enzymes that catalyze DNA synthesis. They add dNTPs to the 3' hydroxyl group of a DNA or RNA primer, using the template sequence to determine which nucleotide to add. The DNA polymerases used in PCR must be able to retain their enzymatic activity after exposure to high temperatures or thermostable. While Taq polymerase was the first thermostable polymerase widely used in PCR, DNA polymerases have been optimized for many different templates and products. For example, if the PCR product will be sequenced a polymerase with 3' to 5' exonuclease "proofreading" activity, which can excise and replace a misincorporated nucleotide, would be a good choice. This type of enzyme may be useful when trying to minimize errors while millions of copies of the template DNA are being made by PCR, which may be important when using the PCR product in downstream applications such as gene expression or DNA sequencing.
Conclusion
PCR can be used to study the vast array of nucleic acid biology. However, along with its many applications, many factors need to be considered when designing a PCR experiment. Some experimental aspects are nearly universal, while others are specific to PCR experiments. Ultimately, the design of a PCR experiment will determine how the PCR product can be analyzed and what information can be learned from it. A great amount of research must be done into a molecular process to design a strong PCR experiment. Watch the “PCR basics” #GenesInSpaceChat on April 20, 2018 on Facebook Live to learn more.